University of Pennsylvania CEIRR (Penn-CEIRR) researchers, Louise Moncla, Ph.D. and Trevor Bedford, Ph.D., built a Nextstrain influenza A/H5N1 cattle outbreak analysis dashboard to investigate the relatedness of the H5N1 viruses infecting cattle to other avian influenza genomes. In collaboration with the Nextstrain team, Dr. Moncla built this analysis using concatenated full genome sequences of H5N1 from cattle and other hosts provided by the U.S. Department of Agriculture (USDA) and GISAID collected during the North American influenza cattle outbreak. In addition, Dr. Moncla and the Nextstrain team have deployed a new visualization for H5 virus evolution that provides an in-depth view of global viral evolution and circulation in the past 2 years. These views allow detailed analysis and investigation of the ongoing outbreak in cattle.
The H5N1 cattle outbreak Nextstrain build is an interactive dashboard to analyze the phylogenetic relationships, transmission events, and nucleotide diversity in avian influenza genomes and includes 218 samples as of May 7th, 2024. The build allows users to customize visualizations by host species, genotype, and “admin division” or U.S. state of origin, among others.
By analyzing the amino acid changes in the H5N1 genome sequences from cattle, Dr. Moncla found a clear branch point where the cluster of cattle influenza sequences diverged from the strain isolated from the single human infection attributed to this outbreak. This branching pattern resulted from a mutation in the influenza PB2 gene that caused a methionine to leucine substitution at position 631 (M631L). Analyzing branch patterns is just one example of this Nextstrain’s capabilities. Dr. Moncla’s Nextstrain build will certainly improve our understanding of the evolution of avian influenza during the cattle outbreak and spark questions for further experimentation.
To learn more about the Nextstrain build, check out this Twitter/X thread from Dr. Moncla. Stay up to date on the cattle outbreak by tracking the virus’s evolution in Nextstrain and follow Dr. Moncla on Twitter/X.
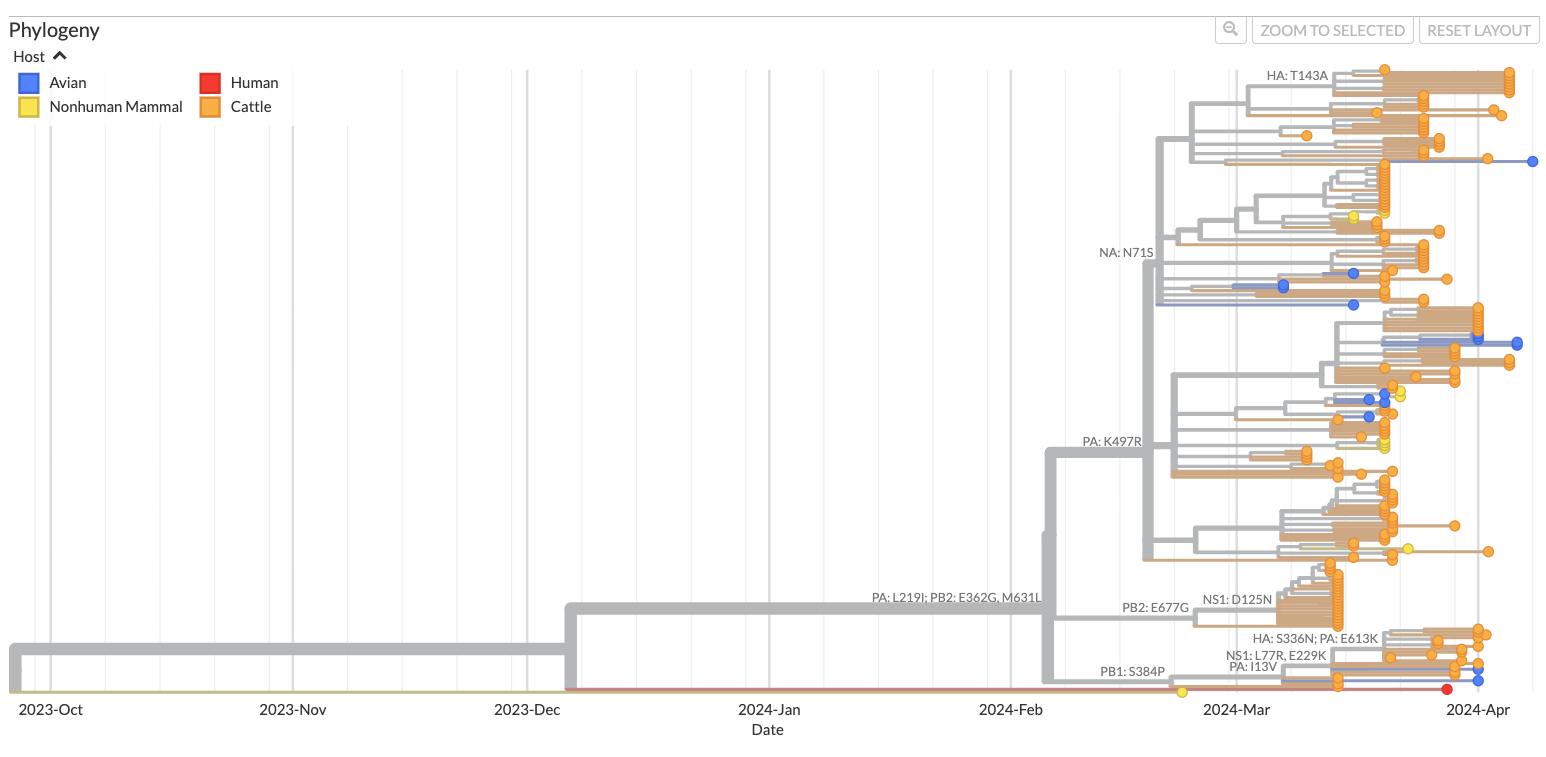 Full genome phylogeny of avian H5N1 from cattle outbreak colored by host species. Represents data in Nextstrain last updated on May 8th, 2024.
Full genome phylogeny of avian H5N1 from cattle outbreak colored by host species. Represents data in Nextstrain last updated on May 8th, 2024.