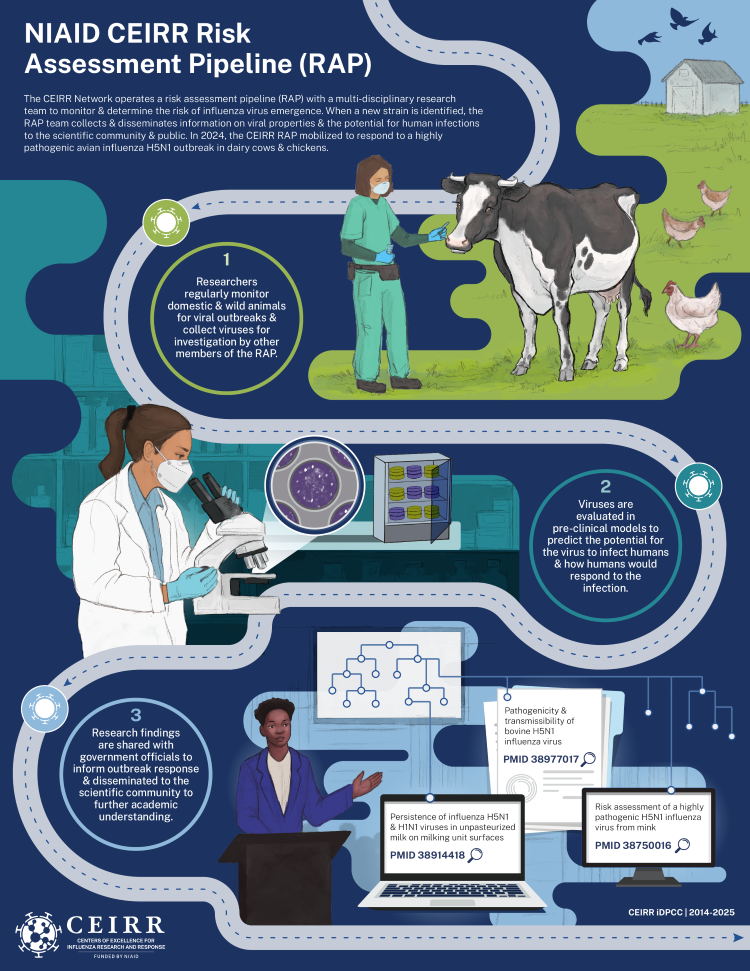
Two of the CEIRR Network’s core missions in studying respiratory viruses with pandemic potential include performing surveillance and responding to outbreaks in wild and domestic animals. In previous infobytes in this series, we discussed the importance of surveilling for influenza viruses in marine mammals and wild birds to understand the virus’s characteristics in nature and to identify strains with the potential to “spillover” into humans and other animals. The CEIRR Network formalized this process of determining whether an influenza strain poses a threat to humans or other animals in what’s called the Risk Assessment Pipeline (RAP). Researchers from various disciplines all collaborate as part of the CEIRR RAP in an effort to minimize the impact posed by emerging viruses. The group identifies viral strains through sampling efforts, tests and characterizes those strains in the lab, and communicates their findings to the scientific community, government officials, and the public. Through this standardized workflow, the CEIRR RAP can evaluate the risk that influenza strains pose to human or animal health and develop strategies to mitigate potential harm. The CEIRR RAP team’s infrastructure allowed them to rapidly mobilize in response to the H5N1 outbreak in dairy cattle. To learn more about the RAP group’s efforts, the iDPCC connected with Anice Lowen, Ph.D., Emory University CEIRR (Emory-CEIRR) PI and leader of the CEIRR RAP, to discuss how she started studying influenza, the strengths and value of the RAP group, and how CEIRR applied the RAP to the bovine H5N1 outbreak.
What Sparked Her Research Into Influenza Biology?
While working toward her biochemistry degree at the University of Alberta, an undergraduate course on viruses immediately captured Dr. Lowen’s interest in virology, and she “never looked back.” She remembers finding these small entities fascinating – wanting to understand how they work, cause disease, and transmit. She began formally studying viruses during graduate school, where she worked on bunyaviruses. When it came time to choose a new focus for her post-doctoral fellowship, she was drawn to influenza. There was a wealth of literature, experimental tools, and expertise in the field, yet so many critical questions about its properties remained unanswered. For a virus that causes such a major burden on public health, Dr. Lowen was motivated to tackle these critical questions to limit the impact of influenza on the population. After settling in Dr. Peter Palese’s lab at the Icahn School of Medicine at Mount Sinai, she set off on her journey to understand influenza biology and the risks it poses to human health.
The Value of CEIRR’s Viral Risk Assessment Pipeline
Dr. Lowen describes viral risk assessment as, “a process of gathering evidence that can help us make a judgment of the likelihood of a pathogen causing an outbreak.” This evidence can come in a variety of forms, which is why the CEIRR RAP group is composed of four sub-groups:
- Domestic Animal Surveillance
- Wild Animal Surveillance
- Phylodynamics and Computational Modeling
- Phenotypic Characterization
Each subgroup studies different aspects of a potential viral threat, such as how and how often a virus spreads, the population’s pre-existing immunity from other strains of the virus, the strain’s mutations and evolutionary patterns, disease severity, and potential treatment options, among other factors. Evaluating all these factors together gives the RAP group a holistic understanding of a virus’s threat level, which they can use to make recommendations for government response and public health efforts.
The risk assessment process is crucial, as Dr. Lowen explains, because “with influenza viruses and other pathogens, they sporadically cause large outbreaks in humans. It's not happening at a regular interval.” Unfortunately, the exact conditions that facilitate outbreaks and pandemics aren’t clear. This means that infectious disease researchers need to stay vigilant to help anticipate these outbreaks and signal when the rest of the population should prepare. These preparations could take different forms, depending on the threat, like making a vaccine, stockpiling antiviral drugs, or changing practices to focus on prevention. By focusing on risk assessment, the CEIRR RAP group can start to find patterns to identify viruses or situations that pose the greatest risk to humans. Then, says Dr. Lowen, “we can have a chance of limiting the likelihood of a pandemic and improving how our population fares during a pandemic.”
Challenges in Viral Risk Assessment
From Dr. Lowen’s perspective, the biggest challenge for influenza risk assessment lies in a fundamental lack of understanding for what allows pandemics to occur. The field learned through decades of research that influenza pandemics are sparked by a virus circulating in a non-human host that “spills over” into humans (can cause infections in humans) and gains the ability to transmit from human to human. However, the specifics remain more elusive.
What types of influenza viruses are most likely to cause a pandemic? Of the four main types of influenza viruses, A, B, C, and D, the influenza A viruses (IAVs) are the only group that have caused pandemics in the past. But within the IAVs, there are dozens of subtypes due to the 18 different hemagglutinin (HA) and 11 different neuraminidase (NA) proteins that could be on the surface of the virus. Another question that Dr. Lowen posed was, “how do [the viruses] need to change in order to transmit among humans?” From past research, the field discovered viral phenotypes that are important to cause a pandemic, like how the viral polymerase adapts or the receptor binding preferences change for a new host. While these phenotypes are necessary for a virus to develop pandemic potential, they aren’t sufficient alone. There are one or more missing pieces that researchers must find to understand the bigger picture.
To find these missing pieces, research focused on discovery is needed. However, as Dr. Lowen notes, “discovery is not fast.” When responding to an outbreak in real time, the field tends to focus on the viral phenotypes that were important in the past to characterize the emerging viral strain. This provides measurable outcomes to evaluate risk but doesn’t advance discovery efforts. Dr. Lowen describes this balancing act as “a bit of a tension between the sort of slower paced research that can give us those really impactful discoveries and the necessary fast paced response to an outbreak.” Luckily with time, these discovery efforts prove fruitful and can improve the fast-paced response efforts for future outbreaks.
The CEIRR RAP Response to the H5N1 Outbreak in Dairy Cattle
Researchers in the CEIRR Network have rich influenza expertise and leveraging that through collaboration bolsters the network’s ability to quickly respond to a potential threat. According to Dr. Lowen, this synergy in the RAP group is what drives the impact. The leaders of the four CEIRR RAP subgroups (outlined above) function as strategists to improve efficiency in risk assessment practices and data/sample flow across the Network. The two surveillance subgroups collect and analyze samples to pass on metadata, viral isolates, and genetic sequence data to the rest of the group. The phylodynamics subgroup utilizes the genetic sequence data to derive insights on viral evolution and transmission and provide epidemiological modeling. The phenotypic characterization subgroup performs experiments to understand whether and how the viruses circulating in nature are changing their phenotypes or risk profiles. The rapid sharing of data across teams with complementary expertise accelerates progress, in the form of new tools and systems, and response efforts.
In March of 2024, the CEIRR RAP group mobilized to respond to the avian influenza H5N1 outbreak in dairy cattle. To rapidly share updates with the scientific community, researchers initially published their results in Bovine H5N1 Outbreak news articles on the CEIRR public website. This outbreak marked the first known spillover of influenza from birds into dairy cattle. Though scientific literature suggested that dairy cattle were susceptible to infection, Dr. Lowen remarked that this was “very surprising for those of us in the field.” This presented a challenge for the CEIRR RAP group because the infrastructure to monitor influenza and respond to outbreaks in dairy cattle didn’t exist.
In contrast to the dairy cattle outbreak, the routine surveillance in swine and poultry offered robust infrastructure to respond to localized influenza outbreaks. The USDA’s surveillance in swine can reveal what strains are circulating and how the strains are changing over time. Additionally, the vaccines administered to swine help to address outbreaks. For poultry, isolation is part of the initial response when influenza is detected. However, infected poultry succumb to disease quite quickly, so flocks need to be culled to prevent influenza spread to additional flocks. Ultimately, the infrastructure and past experience exists in these systems to mitigate risks to humans.
The response mechanisms in dairy cattle are being established in real time, causing problems for not only the CEIRR response, but the national response. Despite this, Dr. Lowen shared that the CEIRR RAP learned and contributed a lot to this outbreak response. Many researchers expanded on their expertise in swine and avian influenza to learn about bovine infections “from the ground up,” and began recruiting bovine experts to the Network. “Some of the really rapid response research that the group has been able to put out there, including experimental evaluation of H5N1 infection in cows,” says Dr. Lowen, “…has been very impressive.”
Dr. Lowen’s lab specifically contributes to the H5N1 outbreak in dairy cattle by studying the fitness of the bovine viruses in the tissues of the human eye. Typically, these viruses cause severe respiratory symptoms. However, conjunctivitis is a prominent feature of the rare human infections by the bovine virus and is new for H5N1 viruses. Additionally, her lab wants to assess the virus’s potential to evolve in the human eye in a way that would increase the risk of the virus spreading to the human respiratory tract or increase its risk of transmission. “In other words,” explains Dr. Lowen, “we don't understand the selective environment of the human eye, and whether it would ‘look’ for viruses that would also replicate better in the human respiratory tract.” She’s hopeful that her lab’s discoveries will inform future risk assessment for zoonosis and pandemics.
To close out our discussion, Dr. Lowen praised the CEIRR RAP group’s efforts, calling it an “excellent job of responding to the present situation to gather data that are really important.” The outcomes from the CEIRR RAP group’s response to the bovine outbreak will inform current and future response efforts and identify prevention strategies to limit spillover in other domestic animals or human infection from occupational exposure. The ongoing discovery research will also aid in the community’s understanding of viral changes that lead to pandemics. On a personal note, Dr. Lowen said she’s “proud to be part of the effort and to have this role in leading it,” and that it was “very gratifying to see the Network step up to the challenge and generate valuable data in a short amount of time.” She credits all her CEIRR colleagues for taking action and working together to respond to the bovine outbreak, because the synergy within this network-wide effort led to its invaluable impact.
To read more about this topic, check out:
- Le Sage, V., Campbell, A. J., Reed, D. S., Duprex, W. P., & Lakdawala, S. S. (2024). Persistence of Influenza H5N1 and H1N1 Viruses in Unpasteurized Milk on Milking Unit Surfaces. Emerging infectious diseases, 30(8), 1721–1723.
- Eisfeld, A. J., Biswas, A., Guan, L., Gu, C., Maemura, T., Trifkovic, S., Wang, T., Babujee, L., Dahn, R., Halfmann, P. J., Barnhardt, T., Neumann, G., Suzuki, Y., Thompson, A., Swinford, A. K., Dimitrov, K. M., Poulsen, K., & Kawaoka, Y. (2024). Pathogenicity and transmissibility of bovine H5N1 influenza virus. Nature, 633(8029), 426–432.
- Restori, K. H., Septer, K. M., Field, C. J., Patel, D. R., VanInsberghe, D., Raghunathan, V., Lowen, A. C., & Sutton, T. C. (2024). Risk assessment of a highly pathogenic H5N1 influenza virus from mink. Nature communications, 15(1), 4112.
- Bovine H5N1 Outbreak news articles on the CEIRR public website to rapidly publish results during the early phase of the outbreak.
- Additional CEIRR publications on H5N1.